Artificial intelligence in hepatopancreaticobiliary surgery: a systematic review
Abstract
Aim: The aim of this systematic review was to provide an overview of Machine Learning applications within hepatopancreaticobiliary surgery. The secondary aim was to evaluate the predictive performances of applied Machine Learning models.
Methods: A systematic search was conducted in PubMed, EMBASE, Cochrane, and Web of Science. Studies were only eligible for inclusion when they described Machine Learning in hepatopancreaticobiliary surgery. The Cochrane and PROBAST risk of bias tools were used to evaluate the quality of studies and included Machine Learning models.
Results: Out of 1821 articles, 52 studies have met the inclusion criteria. The majority of Machine Learning models were developed to predict the course of disease, and postoperative complications. The course of disease has been predicted with accuracies up to 99%, and postoperative complications with accuracies up to 89%. Most studies had a retrospective study design, in which external validation was absent for Machine Learning models.
Conclusion: Machine learning models have shown promising accuracies in the prediction of short-term and long-term surgical outcomes after hepatopancreaticobiliary surgery. External validation of Machine Learning models is required to facilitate the clinical introduction of Machine Learning.
Keywords
INTRODUCTION
Artificial intelligence (AI) has made major progress in healthcare recently, causing an increase in interest in AI algorithms within clinical settings. This may signify the start of a revolutionized digital era within the field of medicine[1].
Artificial intelligence has been defined as the ability of machines to demonstrate human behavior and intelligence[2]. As a major division of AI, Machine Learning (ML) models are able to improve by learning from large-scale data[3]. Decision Tree, Gradient Boosting (GBM), Random Forest, and Support Vector Machine algorithms (SVM) are frequently applied models of ML. A specific branch of ML is known as Deep Learning (DL), which includes multiple layers to recognize several features and patterns from large data[4]. In each layer, values are added to all extracted features. In the end, a model with the best prediction of outcomes is achieved based on training and validation. The accuracy is described as the predictive evaluation of these models on new unseen data. Neural Networks form the basis of DL models and are able to recognize data patterns by using processing layers. Deep Learning functions similarly; however, these models have more layers or depth than Neural Networks. Another group of AI includes Radiomics, which is able to examine various medical images for the purpose of detecting features that are associated with the disease or prognosis[5]. An overview of AI terminology is shown in Table 1.
Definitions AI methods
| General term | |
| Machine Learning (ML) | Machine Learning is an umbrella term and is commonly described as computational techniques that are able to perform complex tasks by analyzing large-scale data[6] |
| Analytical approaches | |
| Decision Tree | Within Decision Tree models, data is divided into smaller nodes and branches. Each node represents a variable, and each branch contains a feature of the variable. Features contain two outcomes such as yes or no. Following one outcome at each feature will eventually form the prediction tree for the desired task. In the end, the smallest tree that optimally fits the data will be produced[7] |
| Gradient Boosting (GBM) | Gradient Boosting models begin with forming a model that fits the data. Afterwards, a consecutive model is constructed that concentrates only on inaccurately predicted aspects of data. Models are then combined to form an improved model. This process is repeated until a final model is established with a minimal error in prediction[8] |
| Random Forest | In a Random Forest model, Decision Trees are present for the desired outcome. Each Decision Tree contains a different prediction path based on the values for the selected variables. By combining all Decision Trees, the final most accurate model will be built[9] |
| Support Vector Machine (SVM) | Support Vector Machines are capable of making predictions by finding the optimal border to classify variables or outcomes in two groups[10] |
| Artificial Neural Networks (ANNs) | Artificial Neural Networks are models in which datasets are analyzed by multiple processing layers. In each layer, features of each data point are extracted to recognize patterns; these features contain weighting factors within each layer. After repeating this training process on multiple datasets, a final model is produced for the complex task[11] |
| Convolutional Neural Networks (CNNs) | Convolutional Neural Networks are similar to ANNs, except these models do not use weights for extracted features. Instead, specific filters are applied to detect patterns in datasets. Additionally, connections are present to provide feedback in each training process[12] |
| Deep Learning | Deep Learning algorithms function similarly to Neural Networks; however, Deep Learning models have more layers or depth than Neural Networks[13] |
| Area of AI that could benefit from ML | |
| Radiomics | In Radiomics models, images are analyzed to detect various quantitative features. Afterwards, these features are used for predictions or associations of several medical outcomes[14] (editors, comment #4) |
In clinical practice, ML has already been used for several purposes, such as diagnosis, treatment decisions, and monitoring of patients[15]. These purposes are especially used in medical specialties that use imaging, such as radiology and pathology. Machine Learning has also been applied in general surgery to improve surgical skill training and predict postoperative outcomes[16]. In hepatopancreatobiliary (HPB) surgery, several clinical challenges are still present, as high frequencies of postoperative complications, such as organ failures, infections, and gastrointestinal tract bleedings, have been reported by surgeons[17]. Additionally, the overall prognosis is poor for malignancies in the hepatobiliary tract and pancreas. For patients with hepatobiliary carcinomas, 5-year survival rates of up to 20% have been reported without upfront surgery, whereas survival rates of 45% have been described with upfront surgery[18]. For borderline-resectable pancreatic carcinomas, 5-year survival rates were discovered to be close to 6% with upfront surgery, although survival rates of 20,5% have been found for patients that have received neoadjuvant chemoradiotherapy (nCRT)[19]. To overcome these clinical challenges, ML models could preoperatively predict disease progression, postoperative complications, and prognosis of patients undergoing HPB surgery. Predicting postoperative complications with ML could provide the opportunity to take prophylactic measures. Furthermore, surgeons could decide between upfront surgery or nCRT based on the predicted response of tumors to nCRT.
Although ML algorithms have shown major potential in HPB surgery, the current status and progress of ML within HPB surgery have not been systematically evaluated in recent literature. However, it is essential to bridge this gap in order to understand the predictive capabilities of ML in HPB surgery properly. Therefore, this systematic review aims to provide a comprehensive overview of ML applications within HPB surgery.
METHODS
Search strategy
Literature was retrieved and systematically reviewed in conformity with the PRISMA guidelines and Cochrane Handbook for Systematic Reviews of Interventions version 6.0. Databases PubMed, Embase.com, Clarivate Analytics/Web of Science Core Collection, and the Wiley/Cochrane Library were used to perform a systematic search. The timeframe within the databases was from inception to the 7th of July 2021. The systematic search was performed by Bektaş M and Burchell GL. The search included keywords and free text terms for (synonyms of) “Machine Learning” combined with (synonyms of) “digestive system surgical procedures”. A comprehensive overview of the search terms per database is available in the supplementary materials [Supplementary Tables 1-4]. The search and protocol of this review were not registered in PROSPERO.
Study selection
During the initial step, articles were included when they described ML within general surgery to secure studies with overlapping content. Subsequently, studies were only qualified if they met the following criteria: (1) describing ML methods within HPB surgery; (2) clinical study; and (3) conducted on adults. Studies were excluded when they: (1) reported on reviews, children, and study abstracts; (2) described regression models; and (3) were not written in English. No specific study design was preferred in the inclusion criteria. Two reviewers (Bektaş M, Costa Pereira J) independently performed the title and abstract screening in conformity with the inclusion and exclusion criteria. Studies were qualified for full-text screening when both reviewers agreed on inclusion. Disagreements were resolved by means of discussion between reviewers, resulting in an agreement.
Risk of bias assessment
The methodological quality assessment of included studies was independently performed by two reviewers (Bektaş M, Costa Pereira J) using the ROBINS-I assessment tool[20]. This tool measures the risk of bias in the domains: confounding, participant selection, intervention classification, deviations from intended interventions, missing outcome data, measurement of outcomes, and selection of reported results. Based on these domains, overall risk of bias is determined for each study. Moreover, the PROBAST risk of bias tool was used to evaluate the quality of ML models within studies[21]. Risk of bias domains included participant selection, predictors, outcomes, and analysis.
Data synthesis and outcome assessment
The following data aspects were independently retrieved from each study by two reviewers (Bektaş M, Costa Pereira J): first author, year, country of research, number of patients, study design, surgical procedure, type of ML, purpose of ML, outcome measurements, and predictive performance. The categorization of studies was based on surgical domains, such as liver, biliary, and pancreatic surgery. Subsequently, accuracies of ML studies were reported within each surgical domain. For each study, the mean accuracy (ACC) and area under the curve (AUC) were calculated to represent the predictive performances of ML models. In addition, descriptive statistics were used to calculate the median and range of accuracies for every ML model.
RESULTS
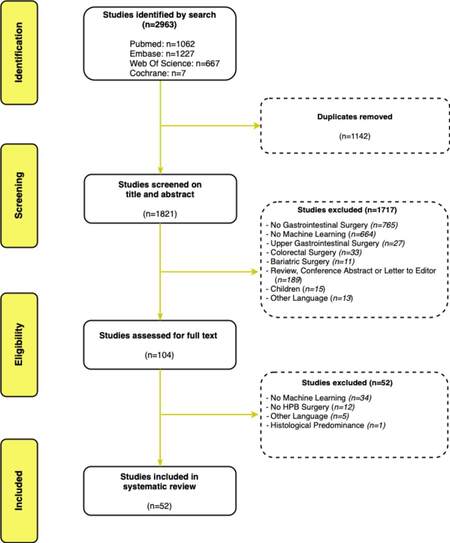
The search strategy identified a total of 1821 studies after the disposal of duplicates [Figure 1]. The 1821 studies were screened for eligibility based on the title and abstract. Subsequently, 104 studies were eligible for full-text assessment, resulting in the inclusion of 52 studies.
Several subclasses of ML have been used within HPB surgery. A vast majority of studies have applied Neural Networks models (n = 16), Radiomics (n = 13), or multiple ML methods (n = 13). Remaining studies involved Decision Trees (n = 7), GBM (n = 1), Random Forest (n = 1), and SVM (n = 1).
Within studies addressing liver surgery, studies predominantly involved hepatocellular carcinomas (HCC)
The purposes of ML algorithms mostly included predicting the course of disease (n = 26), postoperative complications (n = 13), diagnosis (n = 4), and intraoperative complexities (n = 3). Additionally, ML was used to determine essential predictors (n = 5) and to predict the postoperative quality of life (n = 1).
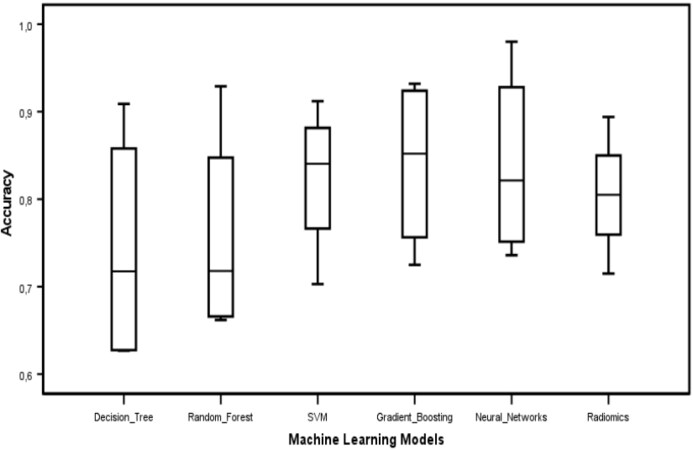
An overview of study characteristics for liver, biliary, and pancreatic surgery is separately presented in Supplementary Tables 5-7, respectively. In addition, the median and range of accuracies for included ML models are presented in Figure 2.
Risk of bias assessment
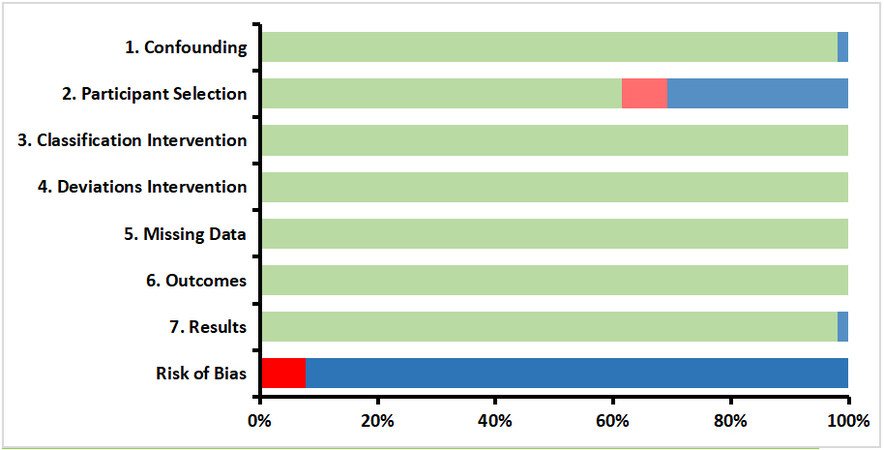
Within the 52 included articles, 47 (90%) retrospective cohort studies have been detected. Additionally, five prospective cohort studies (10%) were present. Therefore, only the ROBINS-I assessment tool was used for the methodological quality assessment. It was discovered that most studies (92%) received a low overall bias, whereas 8% received a high overall bias, mainly due to the presence of selection bias. The results of this assessment are presented in Figure 3. Since we assume that ML models adjust for confounders and are performed consistently, domains such as bias due to confounding and bias in deviations from interventions received low risk of bias scores. However, the bias in the intervention classification domain of these studies received moderate risk of bias scores, because the collection of information occurred before the implementation of ML algorithms.
Figure 3. Methodological quality assessment of the non-randomized studies, according to ROBINS-I assessment tool. Green: Low risk; blue: moderate risk; red: serious risk.
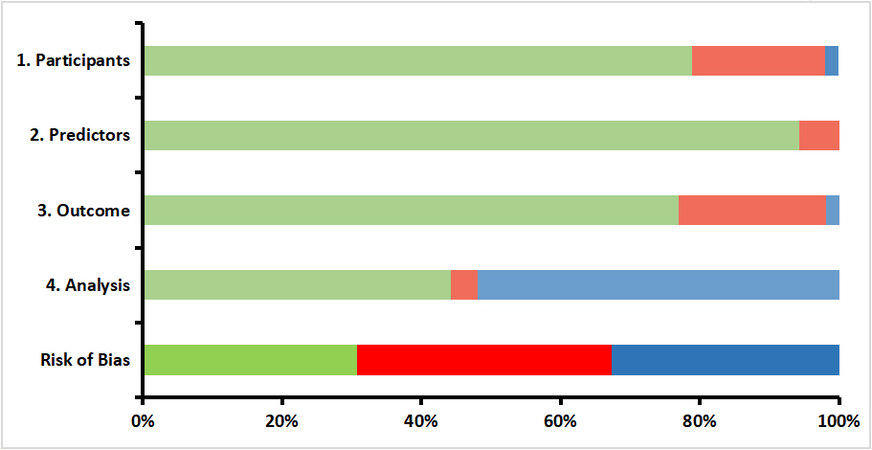
According to the PROBAST risk of bias tool, most studies received a low risk of bias score for the domains selection, predictors, and outcomes. However, the analysis domain, in which missing data and overfitting are accounted for, appeared to have a large proportion of unclear bias scores. Consequently, 30% of the studies received a low overall bias, whereas 33% received an unclear overall bias. A proportion of 37% is covered by studies with an overall high risk of bias, resulting predominantly from bias in the selection and outcomes domains [Figure 4].
Liver surgery
Twenty-one studies have developed ML algorithms to predict the course of disease in patients that underwent hepatectomy for malignancies[22-42]. ML models have shown AUCs between 0.63 and 0.99 for predicting the course of disease, whereas accuracies have been demonstrated to range from 73% to 99%. Eight studies have applied ML to predict postoperative liver function and complications in patients that underwent hepatectomy[43-50]. In predicting postoperative liver function and complications, ML models have demonstrated AUCs ranging from 0.63 to 0.89, and accuracies between 73% and 89% have also been reported. Four studies have used ML to determine predictors and clusters for HCC and ICC patients[51-54]. By using ML, the following significant predictors have been found for the survival of HCC and ICC patients: alpha-fetoprotein, lymphovascular invasion, tumor burden score, tumor number, tumor size, albumin-bilirubin grade, CA 19-9 levels, and neutrophil levels. For CRLM, lymph node metastasis, metastasis size, and carcinoembryonic antigen (CEA) levels appeared to be the key predictors for survival[55].
Biliary surgery
Three studies have developed ML models to predict intraoperative conversions and complexities[56-58]. Intraoperative conversions and complexities have been predicted by ML algorithms with accuracies between 83% and 89%. Two studies applied ML algorithms to predict gallstones and related diseases, in which ML models have shown AUCs from 0.85 to 0.94, along with accuracies up to 97%[59,60]. In addition, Shi et al. applied ML algorithms to predict the postoperative quality of life in patients with gallstones[61]. A mean absolute percentage error of 7.2% and 8.5% was demonstrated, in which a value lower than 10% was considered accurate.
Pancreatic surgery
Five studies have developed ML models to predict the course of disease in patients with pancreas carcinomas who received pancreatectomy procedures[62-66]. AUCs of ML models in predicting the course of disease have ranged from 0.61 to 0.92, and accuracies have been reported to range between 71% and 98%. Additionally, five studies have developed ML algorithms to predict postoperative complications after pancreatic surgery[67-71]. For predicting postoperative complications, ML algorithms have demonstrated AUCs between 0.67 and 0.85, whereas accuracies have varied from 75% to 85%. Two studies have trained ML models to diagnose IPMN in patients that underwent pancreatectomy, in which IPMN’s were diagnosed with AUCs of 0.79 and 0.98[72,73].
DISCUSSION
This review provides an overview of ML applications within HPB surgery. Several ML models have been applied within HPB surgery, in which Neural Networks and Radiomics have been used most frequently. Machine Learning has predominantly been demonstrated for predicting the course of disease, and postoperative complications. Neural Networks have shown the highest predictive performance based on the mean accuracy of 88%. The findings of this study suggest that ML algorithms have promising capacities for patients undergoing HPB surgery.
In predicting the course of disease for patients with HPB malignancies, accuracies of ML models have varied between 61% and 99%. As a comparison, regression models have predicted similar outcomes with accuracies up to 82%[74,75]. For years, HPB surgeons have experienced difficulties in treatment strategies for HPB cancer[76,77]. Multiple clinical trials are conducted to develop optimal treatment strategies to improve patient outcomes after surgery[78]. By using ML to predict metastasis and response to chemotherapy, HPB surgeons could decide to tailor surgery or chemotherapy to patients that could optimally benefit from these treatments.
Machine Learning models have demonstrated accuracies ranging from 63% to 89% for predicting postoperative complications after HPB surgery. Although clinical risk prediction models have been developed to detect postoperative complications, these models have not shown significant improvements compared to the surgeon’s assessment[79]. In addition, conventional regression models have predicted postoperative complications with AUCs up to 0.74[80,81]. Due to its promising predictive performances, ML has illustrated the potential to surpass the surgeon’s assessment and conventional statistics. Ideally, ML models could facilitate the implementation of prophylactic measures and improved patient monitoring based on the predicted complications. This could prevent delayed hospital discharge for patients with severe postoperative complications.
Machine Learning has shown accuracies between 79% and 98% for the diagnosis of HPB pathologies such as gallstones and IPMNs. Meanwhile, logistic regressions have demonstrated accuracies between 73% and 77% in predicting these outcomes[82,83]. As ML seems to be superior in predictive capacities, these models could be used to preoperatively recognize patients with these pathologies, enabling the possibility to track the most important risk factors early and improve patient monitoring. In addition, intraoperative complexities and conversions during laparoscopic cholecystectomies have been predicted by ML models with accuracies up to 89%, whereas logistic regression models have shown accuracies up to 83%[84]. Recently, computer vision models have been developed to locate anatomic landmarks and assess the grading of operative complexities during laparoscopic cholecystectomies[85,86]. Predicting conversions and detecting complexities during operations by using AI models could support intraoperative decision-making and secure optimal patient safety.
For many years, conventional statistical models have been trained to predict surgical outcomes after HPB surgery. Most ML models in this review have shown median AUCs above 0.8, possibly indicating better discriminative abilities than conventional statistics for predicting surgical outcomes after HPB surgery. Furthermore, ML models are able to perform better predictions if the number of input variables is large, whereas conventional statistics function optimally with a few variables[87]. Since clinical databases are complex and usually contain many variables, ML would be preferred for the analysis of clinical data. However, clinicians have been experiencing difficulties in understanding and interpreting ML methods, which is also called the “black-box problem”[88]. This problem could eventually hinder the development and implementation of ML models.
This review has some limitations. Due to inconsistencies in applied ML frequencies, mean accuracies might be underrepresented for a few ML models. Additionally, some of the studies have not reported accuracies or AUCs for the ML algorithm; therefore, a meta-analysis could not be performed.
Since predictive accuracies above 70% indicate good discriminative abilities[89], ML algorithms within this review seem to have promising predictive capabilities for outcomes after HPB surgery. However, as most studies (85%) are missing external validation for the ML algorithms, the generalizability of these models is not supported. The clinical integration of ML could be dependent on this external validation. Therefore, future studies should focus on gaining external validation, which could be facilitated by retrieving large datasets from available patient databases. In addition, interdisciplinary collaborations could be essential in solving this “black-box problem” and support the development of efficient ML models. Data scientists and clinicians should share clinical data to ensure proper data arrangements, data processing, and transparency in methodologies. Sharing data between medical fields might improve the accuracy of ML models and facilitate the procurement of external validation[90].
In conclusion, ML models have shown promising predictive capabilities for relevant clinical challenges and surgical outcomes in HPB surgery. The potential of ML has been demonstrated for pre-, intra-, and postoperative purposes. Therefore, future studies should focus on gaining external validation to facilitate the clinical introduction of ML.
DECLARATIONS
Authors’ contributionsParticipated in the design of the study, data collection and interpretation, wrote and submitted the manuscript: Bektaş M
Revised the manuscript critically and wrote parts of the manuscript: Zonderhuis BM
Revised the manuscript critically and wrote parts of the manuscript: Marquering HA
Participated in the design of the study, and interpretation of data: Costa Pereira J
Performed the literature search: Burchell GL
Participated in the design of the study, and revised the manuscript critically: van der Peet DL
All authors approved the final version of the manuscript.
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2022.
Supplementary MaterialsREFERENCES
1. Briganti G, Le Moine O. Artificial intelligence in medicine: today and tomorrow. Front Med (Lausanne) 2020;7:27.
2. Visvikis D, Cheze Le Rest C, Jaouen V, Hatt M. Artificial intelligence, machine (deep) learning and radio(geno)mics: definitions and nuclear medicine imaging applications. Eur J Nucl Med Mol Imaging 2019;46:2630-7.
3. Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J 2019;6:94-8.
4. Mintz Y, Brodie R. Introduction to artificial intelligence in medicine. Minim Invasive Ther Allied Technol 2019;28:73-81.
6. El Naqa I, Murphy MJ. What is machine learning? In: El Naqa I, Li R, Murphy M, editors. Machine learning in radiation oncology. Cham: Springer; 2015. p. 3-11.
7. Song YY, Lu Y. Decision tree methods: applications for classification and prediction. Shanghai Arch Psychiatry 2015;27:130-5.
8. Friedman JH. Stochastic gradient boosting. Computational Statistics & Data Analysis 2002;38:367-78.
10. Zhang L, Zhou W, Jiao L. Wavelet support vector machine. IEEE Trans Syst Man Cybern B Cybern 2004;34:34-9.
11. Abraham A. Artificial neural networks. Handbook of measuring system design. New Jersey: John Wiley & Sons; 2005. p. 901-8.
14. Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48.
16. Andras I, Mazzone E, van Leeuwen FWB, et al. Artificial intelligence and robotics: a combination that is changing the operating room. World J Urol 2020;38:2359-66.
17. Spolverato G, Ejaz A, Hyder O, Kim Y, Pawlik TM. Failure to rescue as a source of variation in hospital mortality after hepatic surgery. Br J Surg 2014;101:836-46.
18. Pulte D, Weberpals J, Schröder CC, et al. GEKID Cancer Survival Working Group. Survival of patients with hepatobiliary tract and duodenal cancer sites in Germany and the United States in the early 21st century. Int J Cancer 2018;143:324-32.
19. Versteijne E, van Dam JL, Suker M, et al. Dutch Pancreatic Cancer Group. Neoadjuvant chemoradiotherapy versus upfront surgery for resectable and borderline resectable pancreatic cancer: long-term results of the dutch randomized preopanc trial. J Clin Oncol 2022;40:1220-30.
20. Sterne JA, Hernán MA, Reeves BC, et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016;355:i4919.
21. Moons KGM, Wolff RF, Riley RD, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med 2019;170:W1-W33.
22. Mai RY, Zeng J, Meng WD, et al. Artificial neural network model to predict post-hepatectomy early recurrence of hepatocellular carcinoma without macroscopic vascular invasion. BMC Cancer 2021;21:283.
23. Chong H, Gong Y, Pan X, et al. Peritumoral dilation radiomics of gadoxetate disodium-enhanced MRI excellently predicts early recurrence of hepatocellular carcinoma without macrovascular invasion after hepatectomy. J Hepatocell Carcinoma 2021;8:545-63.
24. Ning P, Gao F, Hai J, et al. Application of CT radiomics in prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY) 2020;45:64-72.
25. Shan QY, Hu HT, Feng ST, et al. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging 2019;19:11.
26. Wang W, Chen Q, Iwamoto Y, et al. Deep fusion models of multi-phase CT and selected clinical data for preoperative prediction of early recurrence in hepatocellular carcinoma. IEEE Access 2020;8:139212-20.
27. Ji GW, Zhu FP, Xu Q, et al. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine 2019;50:156-65.
28. Qin H, Hu X, Zhang J, et al. Machine-learning radiomics to predict early recurrence in perihilar cholangiocarcinoma after curative resection. Liver Int 2021;41:837-50.
29. Schoenberg MB, Bucher JN, Koch D, et al. A novel machine learning algorithm to predict disease free survival after resection of hepatocellular carcinoma. Ann Transl Med 2020;8:434.
30. Chiu HC, Ho TW, Lee KT, Chen HY, Ho WH. Mortality predicted accuracy for hepatocellular carcinoma patients with hepatic resection using artificial neural network. ScientificWorldJournal 2013;2013:201976.
31. Qiao G, Li J, Huang A, Yan Z, Lau WY, Shen F. Artificial neural networking model for the prediction of post-hepatectomy survival of patients with early hepatocellular carcinoma. J Gastroenterol Hepatol 2014;29:2014-20.
32. Spelt L, Nilsson J, Andersson R, Andersson B. Artificial neural networks - a method for prediction of survival following liver resection for colorectal cancer metastases. Eur J Surg Oncol 2013;39:648-54.
33. Ho WH, Lee KT, Chen HY, Ho TW, Chiu HC. Disease-free survival after hepatic resection in hepatocellular carcinoma patients: a prediction approach using artificial neural network. PLoS One 2012;7:e29179.
34. Dong Y, Zhou L, Xia W, et al. Preoperative prediction of microvascular invasion in hepatocellular carcinoma: initial application of a radiomic algorithm based on grayscale ultrasound images. Front Oncol 2020;10:353.
35. Feng ST, Jia Y, Liao B, et al. Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol 2019;29:4648-59.
36. Song D, Wang Y, Wang W, et al. Using deep learning to predict microvascular invasion in hepatocellular carcinoma based on dynamic contrast-enhanced MRI combined with clinical parameters. J Cancer Res Clin Oncol 2021;147:3757-67.
37. Zhou W, Jian W, Cen X, et al. Prediction of microvascular invasion of hepatocellular carcinoma based on contrast-enhanced MR and 3D convolutional neural networks. Front Oncol 2021;11:588010.
38. Mao B, Ma J, Duan S, Xia Y, Tao Y, Zhang L. Preoperative classification of primary and metastatic liver cancer via machine learning-based ultrasound radiomics. Eur Radiol 2021;31:4576-86.
39. Yao X, Huang X, Yang C, et al. A novel approach to assessing differentiation degree and lymph node metastasis of extrahepatic cholangiocarcinoma: prediction using a radiomics-based particle swarm optimization and support vector machine model. JMIR Med Inform 2020;8:e23578.
40. Sahara K, Paredes AZ, Tsilimigras DI, et al. Machine learning predicts unpredicted deaths with high accuracy following hepatopancreatic surgery. Hepatobiliary Surg Nutr 2021;10:20-30.
41. Hamamoto I, Okada S, Hashimoto T, Wakabayashi H, Maeba T, Maeta H. Prediction of the early prognosis of the hepatectomized patient with hepatocellular carcinoma with a neural network. Computers in Biology and Medicine 1995;25:49-59.
42. Zhu HB, Xu D, Ye M, et al. Deep learning-assisted magnetic resonance imaging prediction of tumor response to chemotherapy in patients with colorectal liver metastases. Int J Cancer 2021;148:1717-30.
43. Chen Y, Liu Z, Mo Y, et al. Prediction of post-hepatectomy liver failure in patients with hepatocellular carcinoma based on radiomics using Gd-EOB-DTPA-enhanced MRI: the liver failure model. Front Oncol 2021;11:605296.
44. Zhu WS, Shi SY, Yang ZH, Song C, Shen J. Radiomics model based on preoperative gadoxetic acid-enhanced MRI for predicting liver failure. World J Gastroenterol 2020;26:1208-20.
45. Mai RY, Lu HZ, Bai T, et al. Artificial neural network model for preoperative prediction of severe liver failure after hemihepatectomy in patients with hepatocellular carcinoma. Surgery 2020;168:643-52.
46. Mai RY, Zeng J, Mo YS, et al. Artificial neural network model for liver cirrhosis diagnosis in patients with hepatitis B virus-related hepatocellular carcinoma. Ther Clin Risk Manag 2020;16:639-49.
47. Kato H, Kanematsu M, Zhang X, et al. Computer-aided diagnosis of hepatic fibrosis: preliminary evaluation of MRI texture analysis using the finite difference method and an artificial neural network. AJR Am J Roentgenol 2007;189:117-22.
48. Zhang T, Wei Y, He X, et al. Prediction of remnant liver regeneration after right hepatectomy in patients with hepatocellular carcinoma using preoperative CT texture analysis and clinical features. Contrast Media Mol Imaging 2021;2021:5572470.
49. Merath K, Hyer JM, Mehta R, et al. Use of machine learning for prediction of patient risk of postoperative complications after liver, pancreatic, and colorectal surgery. J Gastrointest Surg 2020;24:1843-51.
50. Lei L, Wang Y, Xue Q, Tong J, Zhou CM, Yang JJ. A comparative study of machine learning algorithms for predicting acute kidney injury after liver cancer resection. PeerJ 2020;8:e8583.
51. Tsilimigras DI, Mehta R, Moris D, et al. Utilizing machine learning for pre- and postoperative assessment of patients undergoing resection for BCLC-0, A and B hepatocellular carcinoma: implications for resection beyond the BCLC guidelines. Ann Surg Oncol 2020;27:866-74.
52. Tsilimigras DI, Mehta R, Moris D, et al. A machine-based approach to preoperatively identify patients with the most and least benefit associated with resection for intrahepatic cholangiocarcinoma: an international multi-institutional analysis of 1146 patients. Ann Surg Oncol 2020;27:1110-9.
53. Bagante F, Spolverato G, Merath K, et al. Intrahepatic cholangiocarcinoma tumor burden: a classification and regression tree model to define prognostic groups after resection. Surgery 2019;166:983-90.
54. Tsilimigras DI, Hyer JM, Paredes AZ, et al. A novel classification of intrahepatic cholangiocarcinoma phenotypes using machine learning techniques: an international multi-institutional analysis. Ann Surg Oncol 2020;27:5224-32.
55. Moro A, Mehta R, Tsilimigras DI, et al. Prognostic factors differ according to KRAS mutational status: a classification and regression tree model to define prognostic groups after hepatectomy for colorectal liver metastasis. Surgery 2020;168:497-503.
56. Gholipour C, Fakhree MB, Shalchi RA, Abbasi M. Prediction of conversion of laparoscopic cholecystectomy to open surgery with artificial neural networks. BMC Surg 2009;9:13.
57. Eldar S, Siegelmann HT, Buzaglo D, et al. Conversion of laparoscopic cholecystectomy to open cholecystectomy in acute cholecystitis: artificial neural networks improve the prediction of conversion. World J Surg 2002;26:79-85.
58. Bouarfa L, Schneider A, Feussner H, et al. Prediction of intraoperative complexity from preoperative patient data for laparoscopic cholecystectomy. Artif Intell Med 2011;52:169-76.
59. Liew PL, Lee YC, Lin YC, et al. Comparison of artificial neural networks with logistic regression in prediction of gallbladder disease among obese patients. Dig Liver Dis 2007;39:356-62.
60. Vukicevic AM, Stojadinovic M, Radovic M, et al. Automated development of artificial neural networks for clinical purposes: Application for predicting the outcome of choledocholithiasis surgery. Comput Biol Med 2016;75:80-9.
61. Shi HY, Lee HH, Tsai JT, et al. Comparisons of prediction models of quality of life after laparoscopic cholecystectomy: a longitudinal prospective study. PLoS One 2012;7:e51285.
62. Velez-Serrano JF, Velez-Serrano D, Hernandez-Barrera V, et al. Prediction of in-hospital mortality after pancreatic resection in pancreatic cancer patients: a boosting approach via a population-based study using health administrative data. PLoS One 2017;12:e0178757.
63. Mofidi R, Duff MD, Madhavan KK, Garden OJ, Parks RW. Identification of severe acute pancreatitis using an artificial neural network. Surgery 2007;141:59-66.
64. Ansari D, Nilsson J, Andersson R, Regnér S, Tingstedt B, Andersson B. Artificial neural networks predict survival from pancreatic cancer after radical surgery. Am J Surg 2013;205:1-7.
65. Walczak S, Velanovich V. An evaluation of artificial neural networks in predicting pancreatic cancer survival. J Gastrointest Surg 2017;21:1606-12.
66. Sala Elarre P, Oyaga-Iriarte E, Yu KH, et al. Use of machine-learning algorithms in intensified preoperative therapy of pancreatic cancer to predict individual risk of relapse. Cancers (Basel) 2019;11:606.
67. Mu W, Liu C, Gao F, et al. Prediction of clinically relevant pancreatico-enteric anastomotic fistulas after pancreatoduodenectomy using deep learning of preoperative computed tomography. Theranostics 2020;10:9779-88.
68. Lin Z, Tang B, Cai J, et al. Preoperative prediction of clinically relevant postoperative pancreatic fistula after pancreaticoduodenectomy. Eur J Radiol 2021;139:109693.
69. Han IW, Cho K, Ryu Y, et al. Risk prediction platform for pancreatic fistula after pancreatoduodenectomy using artificial intelligence. World J Gastroenterol 2020;26:4453-64.
70. Skawran SM, Kambakamba P, Baessler B, et al. Can magnetic resonance imaging radiomics of the pancreas predict postoperative pancreatic fistula? Eur J Radiol 2021;140:109733.
71. Cos H, Li D, Williams G, et al. Predicting outcomes in patients undergoing pancreatectomy using wearable technology and machine learning: prospective cohort study. J Med Internet Res 2021;23:e23595.
72. Kuwahara T, Hara K, Mizuno N, et al. Usefulness of deep learning analysis for the diagnosis of malignancy in intraductal papillary mucinous neoplasms of the pancreas. Clin Transl Gastroenterol 2019;10:1-8.
73. Attiyeh MA, Chakraborty J, Gazit L, et al. Preoperative risk prediction for intraductal papillary mucinous neoplasms by quantitative CT image analysis. HPB (Oxford) 2019;21:212-8.
74. Chen Y, Liu H, Zhang J, et al. Prognostic value and predication model of microvascular invasion in patients with intrahepatic cholangiocarcinoma: a multicenter study from China. BMC Cancer 2021;21:1299.
75. Ren Z, He S, Fan X, et al. Survival prediction model for postoperative hepatocellular carcinoma patients. Medicine (Baltimore) 2017;96:e7902.
76. Orcutt ST, Anaya DA. Liver resection and surgical strategies for management of primary liver cancer. Cancer Control 2018;25:1073274817744621.
78. Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer 2020;1873:188314.
79. Samim M, Mungroop TH, AbuHilal M, et al. HPB-RISC Study Group. Surgeons’ assessment versus risk models for predicting complications of hepato-pancreato-biliary surgery (HPB-RISC): a multicenter prospective cohort study. HPB (Oxford) 2018;20:809-14.
80. Li B, Qin Y, Qiu Z, Ji J, Jiang X. A cohort study of hepatectomy-related complications and prediction model for postoperative liver failure after major liver resection in 1441 patients without obstructive jaundice. Ann Transl Med 2021;9:305.
81. Ma KW, Cheung TT, She WH, et al. Risk prediction model for major complication after hepatectomy for malignant tumour - a validated scoring system from a university center. Surg Oncol 2017;26:446-52.
82. Lu JH, Tong GX, Hu XY, Guo RF, Wang S. Construction and evaluation of a nomogram to predict gallstone disease based on body composition. Int J Gen Med 2022;15:5947-56.
83. Shimizu Y, Hijioka S, Hirono S, et al. New model for predicting malignancy in patients with intraductal papillary mucinous neoplasm. Ann Surg 2020;272:155-62.
84. Kim MS, Kwon HJ, Park HW, et al. Preoperative prediction model for conversion of laparoscopic to open cholecystectomy in patient with acute cholecystitis: based on clinical, laboratory, and CT parameters. J Comput Assist Tomogr 2014;38:727-32.
85. Liu R, An J, Wang Z, et al. Artificial intelligence in laparoscopic cholecystectomy: does computer vision outperform human vision? Art Int Surg 2022;2:80-92.
86. Tranter-entwistle I, Eglinton T, Connor S, Hugh TJ. Operative difficulty in laparoscopic cholecystectomy: considering the role of machine learning platforms in clinical practice. Art Int Surg 2022; doi: 10.20517/ais.2022.01.
88. Grant L, Joo P, Nemnom MJ, Thiruganasambandamoorthy V. Machine learning versus traditional methods for the development of risk stratification scores: a case study using original Canadian Syncope Risk Score data. Intern Emerg Med 2022;17:1145-53.
89. Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 2010;5:1315-6.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Bektaş M, Zonderhuis BM, Marquering HA, Costa Pereira J, Burchell GL, van der Peet DL. Artificial intelligence in hepatopancreaticobiliary surgery: a systematic review. Art Int Surg 2022;2:132-43. http://dx.doi.org/10.20517/ais.2022.20
AMA Style
Bektaş M, Zonderhuis BM, Marquering HA, Costa Pereira J, Burchell GL, van der Peet DL. Artificial intelligence in hepatopancreaticobiliary surgery: a systematic review. Artificial Intelligence Surgery. 2022; 2(3): 132-43. http://dx.doi.org/10.20517/ais.2022.20
Chicago/Turabian Style
Bektaş, Mustafa, Babs M. Zonderhuis, Henk A. Marquering, Jaime Costa Pereira, George L. Burchell, Donald L. van der Peet. 2022. "Artificial intelligence in hepatopancreaticobiliary surgery: a systematic review" Artificial Intelligence Surgery. 2, no.3: 132-43. http://dx.doi.org/10.20517/ais.2022.20
ACS Style
Bektaş, M.; Zonderhuis BM.; Marquering HA.; Costa Pereira J.; Burchell GL.; van der Peet DL. Artificial intelligence in hepatopancreaticobiliary surgery: a systematic review. Art. Int. Surg. 2022, 2, 132-43. http://dx.doi.org/10.20517/ais.2022.20
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 14 clicks
Cite This Article 14 clicks












Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.